Description
Project for a bioinformatics course taken at Sapienza. The task was to find a list of genes that are differentially expressed between case and control samples and characterise them. I used R to analyse a dataset of endometrium and endometriosis samples from GEO to identify differentially expressed genes which could be used for diagnosis and performed functional enrichment analysis on these genes to investigate the disease's pathogenesis.
Inspiration
I chose to analyse endometriosis because it is an under-researched disease. However, recent advancements in genetic sequencing and bioinformatics are promising for new research.
Process
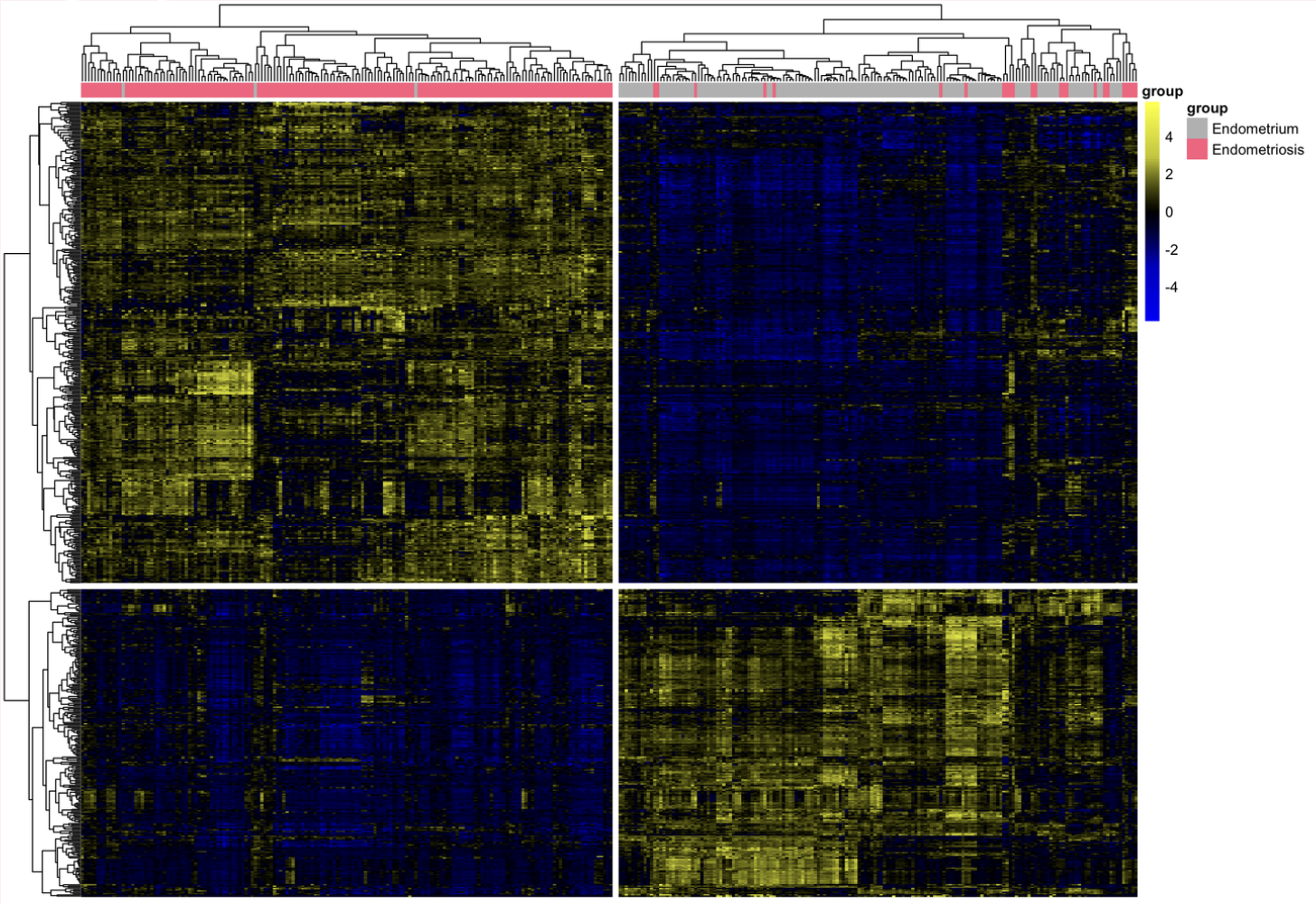
I downloaded the dataset from GEO and performed pre-processing and filtering to keep only the genes that are up or down-regulated by at least double or half.
Next, I performed PCA and clustering on these differentially expressed genes, which confirmed that there was clear linear separation with two principal components and two clear clusters. I did a literature review on the most up and down-regulated genes (SFRP2 and ELAPOR1) and found that both have been proposed as potential diagnostic biomarkers.
Finally, I performed a functional enrichment analysis on the differentially expressed genes using DisGeNet, GO, and KEGG, and did further literature reviews on the most enriched genes. Many of these genes are related to the immune response and recent endometriosis research is heading in this direction.
Learnings
Although there were clear differentially expressed genes, more research is needed to determine whether the proposed diagnostic biomarkers are feasible. The results of the functional enrichment analysis were challenging to navigate and confirmed that the pathogenesis of endometriosis is very complex. Overall, I learnt how to use different biological data sources and tools in bioinformatics.